Hi,
I am the author this application, It will be great that you could also share 15 structures with me once you are OK.
Thanks
Jiazhou
Hi,
I am the author this application, It will be great that you could also share 15 structures with me once you are OK.
Thanks
Jiazhou
Hi Yosukemino,
Thanks for coming up with beta-methyl issue, In the application, 'there is a requirement for the chain to be branched at the position’ this rule is not counted since it’s not included in the guidance. If this rule is something that should be considered first when to come to count a beta-methyl, I can actually fix it in the application.
Thanks
Jiazhou
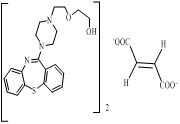
This is the API (Quetipine fumarate) and identified impurities are mentioned below
and i considered these impurities are NDSRIs . Is it correct?>???
![]()
![]()
Request for ur expert opinion on above question ??
Yes, I’ve also tried it. Looks really promising! Thanks a lot for sharing.
Yes, all of them could be potential nitrosamines, although the last one looks the least likely
A similar tool posted by @diullio on CPCA calculator on Python Desktop
Previously available as a preprint and not yet subjected to peer review, the work of Jiazhou Zhu, Qu, and Ye (2024) has now been published.
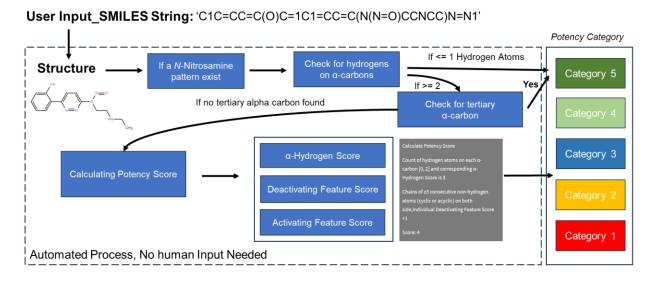
The algorithm quickly and accurately categorized the risks of over 6000 chemicals, with high agreement compared to the dataset labels.
Further analysis revealed a correlation between high pKaH values of the parent amine (the basicity of an amine where the susceptibility to nitrosation is related to the nitrogen basicity of the amine) and low risk of N-nitrosamine.
To ensure the algorithm’s robustness and reliability, testing was conducted on a larger dataset containing NDSRIs. This dataset was obtained from the referenced article:
The Landscape of Potential Small and Drug Substance Related Nitrosamines in Pharmaceuticals
The algorithm accurately calculated the potency category for each chemical within seconds, while also identifying that 16 molecules failed due to an invalid SMILES string.
Since the posting date of the preprint in this community until now, with the article already published, for those who have been using the tool, what new recommendations do you have?
It is very useful . I have applied in many application and found suitable
Dear @Yosukemino & @romnaiffer,
While calculating the CPCA score for 7-nitroso-3-(trifluoromethyl)-5,6,7,8-tetrahydro-[1,2,4]triazolo[4,3- a]pyrazine (NTTP of Sitagliptin) in the CPCA web-based calculator, i am getting AI= 100ng/Day (Score-2), At the same time EMA published “EMA/315970/2023 rev.1” it is showing 37 ng/Day. I am confused to which limit should i choose? Can somebody through light on this for my better understanding.
dear Naveen
The AI of 37ng/day had been established applying the SAR approach using the structural surrogate NTPH (TD50=37ug/kg/day). Last year, EMA published an approach for assigning an N-nitrosamine impurity (including nitrosamine drug substance-related impurities [NDSRIs]) to a predicted carcinogenic potency category, with a corresponding acceptable intake (AI) limit, based on an assessment of activating or deactivating structural features present in the molecule, also known as Carcinogenic Potency Categorization Approach (CPCA). Thus, according to CPCA, NTTP belongs to Category 2 and therefore, a higher AI of 100 ng/day established. please refer to the last update of EMA Appendix 1
Thank you Dear Elenipoliti for the brief clarity.
Hello Sir.
Akshayraj Here.
n-nitroso n-desmethyl ponatinib having piperazine group as shown in "n-nitroso-piperazine. So, what could be the potency score and AI limit for n-nitroso n-desmethyl ponatinib ?
Thanks.
Hi,
Use below CPCA calculation for N-Nitroso N-Desmethyl Ponatinib Impurity
Calculate Potency Score
Count of hydrogen atoms on each α-carbon [2, 2] and corresponding α-Hydrogen Score is 1
Ring related is found, Individual Deactivating Feature Score is +2
Score: 3
Potency Category 3 : AI = 400 ng/day