An intriguing and thought-provoking question arises from the information presented in the Journal Pre-Proof “Acceptable Intakes (AIs) for 11 small molecule N-nitrosamines (NAs)” @jbercu. How can the reliance on the Lhasa Carcinogenicity Database (CDB) and read-across methodologies be enhanced to accommodate the assessment of novel chemical spaces, such as NA drug-substance-related impurities (NDSRIs), which may have distinct metabolic pathways and molecular characteristics?
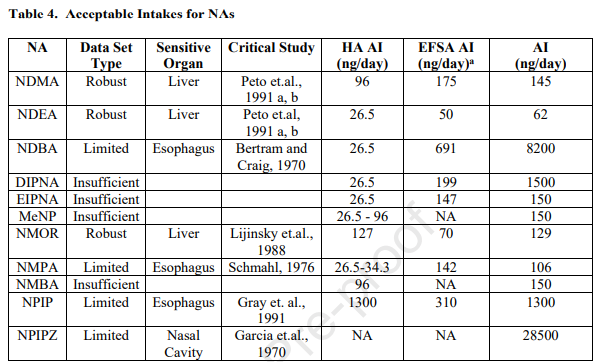
The paper emphasizes the importance of reviewing the literature beyond the Lhasa CDB and highlights that not all studies are captured in this database. This raises concerns regarding the potential gaps in knowledge and the need for toxicologists to seek additional carcinogenicity data. The example of NDEA, with robust dose-response mutagenicity and carcinogenicity studies in rodents, further underscores the significance of comprehensive data analysis.
The revised NDEA acceptable intake (AI) of 62 ng/day, considering the most sensitive site and tumor type, suggests a downstream impact on other limits. This prompts us to question the potential consequences of adjusting the AI for NDEA on other AIs that have been established based on read-across to this compound.
Furthermore, the paper raises an important point regarding the use of more mechanistic and metabolism data in read-across methodologies, particularly in the context of defining AIs for NDSRIs. These impurities often belong to a novel chemical space and exhibit different molecular characteristics and metabolic behaviors compared to small molecule NAs. The current default AI of 18 ng/day for NDSRIs is based on small molecule NAs that are readily metabolized into toxic species. However, the larger molecular weight and unique chemistry of NDSRIs may impede metabolism around the alpha-carbon position, necessitating a re-evaluation of the default AI.
Consequently, a compelling question emerges: How can the Lhasa CDB and read-across methodologies be augmented to effectively incorporate mechanistic and metabolism data, enabling accurate and reliable assessment of carcinogenicity risks associated with novel chemical spaces like NDSRIs?
Addressing this question calls for the exploration of innovative approaches, such as the integration of predictive modeling, in vitro assays, and computational toxicology tools, to enhance the prediction of carcinogenicity and establish appropriate AI limits for NDSRIs. Additionally, it necessitates collaborative efforts among researchers, toxicologists, and regulatory authorities to ensure the utilization of the most up-to-date and comprehensive data sources, enabling a more thorough and scientifically robust evaluation of the potential risks posed by emerging chemical entities.