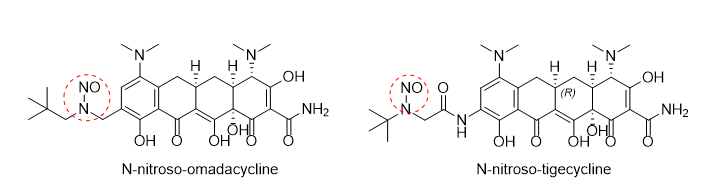
N-nitroso-tigecycline and N-nitroso-omadacycline have similar structurs,but have different AI:N-nitroso-tigecycline is 1500ng/d,N-nitroso-omadacycline is 26.5ng/d.I don’t have a suitable analysis method,How should I do it?
Hi,
The structure of the nitrosamines are different and that could the reason for differences in AI values.
N-nitroso-tigecycline: Has a tertiary carbon in the alpha position.
N-nitroso-Omadacycline: Has an activating aryl group bonded to the alpha carbon.
I have learned,thank you!
EMA has been reported N-nitroso-tigecycline negative in in vivo mutagenicity study.
Is there any literature reporting N-Nitroso Omadacycline mutagenicity?
Read-across based on in vivo mutagenicity data is not unprecedented in the EMA Appendix 1.
In my view, you can consider comparative studies on N-nitroso-tigecycline (or another candidate with data) and N-nitroso-omadacycline to assess whether they are likely of comparable potency.
This research would then address whether the overall bulkiness of the molecules is more determinative for their potency than the local environment around the nitroso bond (aka relevance of benzylic feature). If this is the case, a read-across case would make sense.
You could look for both molecules at physicochemical properties comparison, CYP-NDSRI binding via molecular docking and QM reaction pathway modeling, and use that info to build a hypothesis that can be validated with comparative in vitro (general or CYP-specific) mammalian cell assays or (general or CYP-specific) metabolization studies, maybe doing a comparative in vivo comet assay as well. (The most suitable validation strategy will depend on the data generated.) This seems to me the easiest path to read-across instead of a doing a new in vivo TGR study, whereas Ames testing is not relevant for antibiotics.
If you do studies, please consider publishing as these are key examples for the molecular weight discussion or CPCA refinement and the ICH M7 addendum.
Initially, a QSAR analysis can be performed,but it is expected that the toxicity will not be comparable due to the chemical environment of both structures. Omadacycline has two protons on both alpha carbons, while tigecycline has only one alpha carbon. In addition, omadacycline has an aryl ring attached to the alpha carbon (activating feature), while tigecycline has a carbonyl group (as part of the amide group) attached to the alpha carbon.
Currently, there is no information in the Lhasa carcinogenicity database, so you can perform tests (e.g. comet assay) as mentioned by @ccdw in the previous message.
I do not agree that you can already conclude that the toxicity will not be sufficiently comparable due to the local environment differences, just by looking at the structure.
You first need to know if local environment is more determinative of the risk than the overall MW and bulkiness of the molecule (impacting CYP binding options).
Molecular size does have an impact on CYP activity and thus needs to be considered in the assessment (Kostal 2025). While CPCA overlooks MW (both from the 1 mole of NDSRI, 1 mole of alkylating species principle (Fine 2023; Nudelman 2023) and the influence of MW (or overall bulkiness) on Phase I metabolism options (Kostal 2025, Bercu 2024)), read-across doesn’t necessarily have to.
Indeed, due to the local similarity differences I suggested in my earlier post to assess first “if the overall bulkiness of the molecules is more determinative for their potency than the local environment around the nitroso bond”. For the 2 possible alpha-hydroxylation positions of N-nitroso-OMA and the single possible alpha-hydroxylation position of N-nitroso-TIGE you can evaluate with CYP-NDSRI binding studies via molecular docking if the catalysis is likely, and if some scenarios are sufficiently likely you can supplement this with QM reaction pathway modeling or use these insights to design follow-up studies (unfortunately a comparative Ames test is not an easy follow-up due to to antibiotic activity). (You can also invent structures “in between” OMA and TIGE/substitution variants for computational comparison and increased understanding.)
Note that CPCA takes into account the intrinsic lability of a benzylic C-H and thus its chemical susceptibility to a-hydroxylation and not the likelihood of it leading to a more stabilized carbocation linked to the benzylic substitution or its link with further metabolization step. So, when you address the likelihood of a-hydroxylation in the assessment with different techniques, you address the warnings on this feature. (If a hydrogen is labile but not accessible, the lability will not necessarily be expressed.)
The bulkiness in the local environment is also tackled in the CYP-NDSRI binding studies, whereas the bulkiness of a potential DNA reactive species (if a mechanistic risk for generation is seen) and its influence of reactivity with DNA can be studied in comparative assays, but of course only matters in the next step if Phase I metabolism is evaluated to be sufficiently likely to pose a risk.